» A summary of what happened.
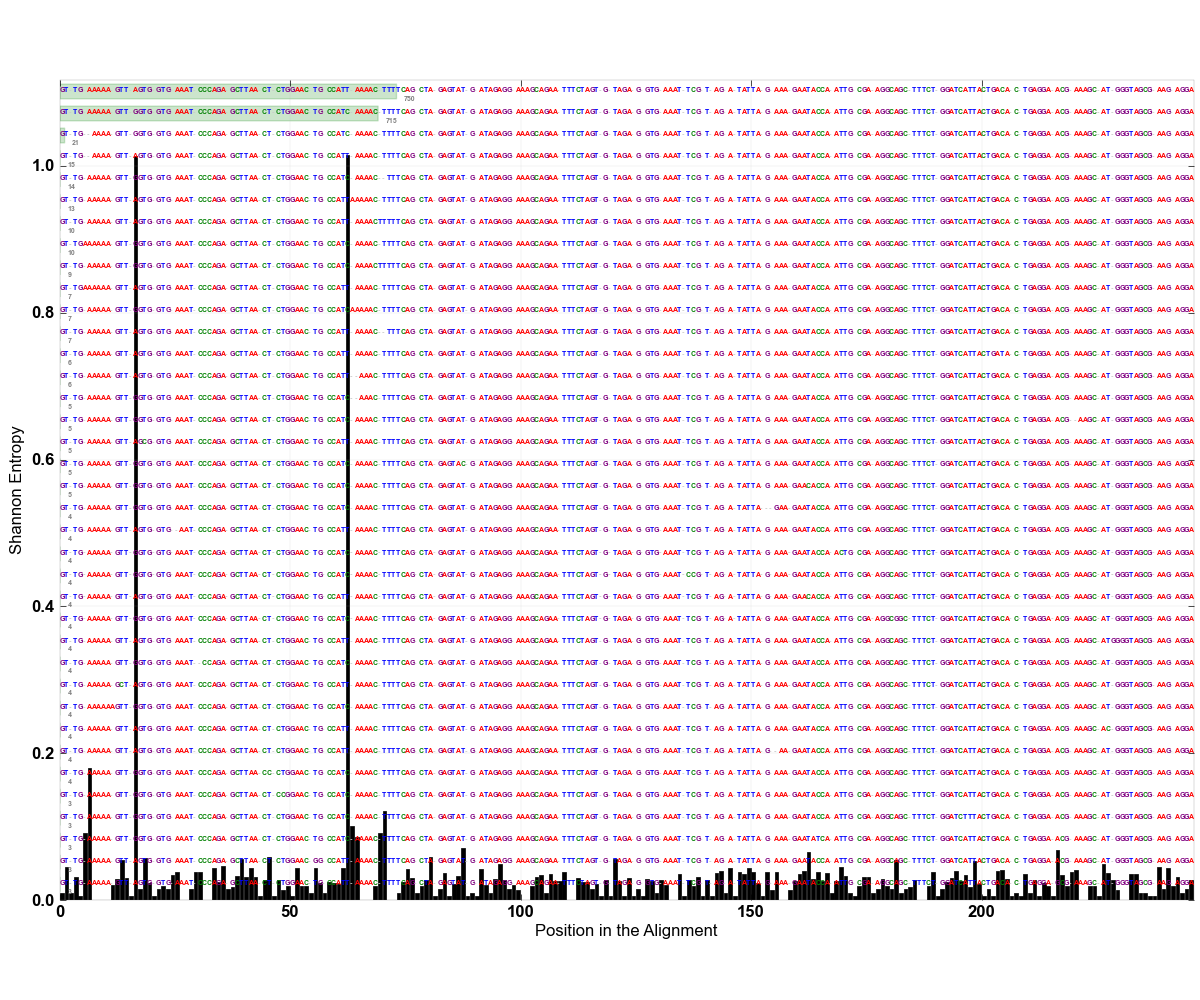
Oligotyping analysis was performed on 2,500 reads from 10 samples for "mock-env-aligned" with oligotyping pipeline version 0.96 (available from http://oligotyping.org) using 2 components automatically selected form the highest entropy values following the initial entropy analysis. To reduce the noise, each oligotype required to (1) appear in at least 1 sample, (2) occur in more than 1.0% of the reads for at least one sample, (3) represent a minimum of 0 reads in all samples combined, and (4) have a most abundant unique sequence with a minimum abundance of 0. Oligotypes that did not meet these criteria were removed from the analysis. The final number of quality controlled oligotypes revealed by the analysis was , and they represented 2478 reads, which was equivalent to 99.12% of all reads analyzed.
» Meta
| Run date | 26 Jul 13 13:45:28 |
| Library version | 0.96 |
» Given Parameters
| Number of entropy components to be selected automatically | 2 |
| Min number of samples oligotype expected to appear | 1 |
| Min % abundance of oligotype in at least one sample | 1.0% |
| Min total abundance of oligotype in all samples | 0 |
| Min substantive abundance of an oligotype (-M) | 0 |
| Quality scores were provided | False |
| Oligotype sets were requested to be generated | False |
» Components used for Oligotyping
| Base locations of interest in the alignment | 16, 62 |
» Quality filtering results
| Number of sequences analyzed | 2,500 |
| Number of unique oligotypes (raw) | 8 |
| Oligotypes after "min number of samples" elimination | 8 |
| Oligotypes after "min % abundance in a sample" elimination | 2 |
| Number of sequences analyzed | 2,500 |
| Number of samples found | 10 |
| Number of sequences represented after quality filtering | 2,478 |
| Percentage of reads represented in results | 99.12% |
» Files to analyze results further via third partry applications
| Representative sequences per oligotype | oligo-representatives.fa.txt |
| Sample/oligotype abundance data matrix (percents) | matrix_percents.txt |
| Sample/oligotype abundance data matrix (counts) | matrix_counts.txt |
| Read distribution among samples table | read_distribution.txt |
| GEXF file for network analysis | network.gexf |
| Environment file | environment.txt |
| FASTA file for abundant oligotypes | oligos.fa.txt |
| NEXUS file for abundant oligotypes | oligos.nex.txt |
» Entropy values that components were picked from
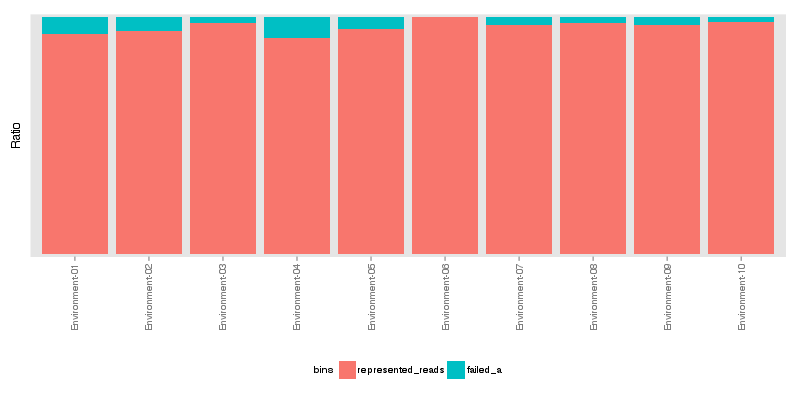
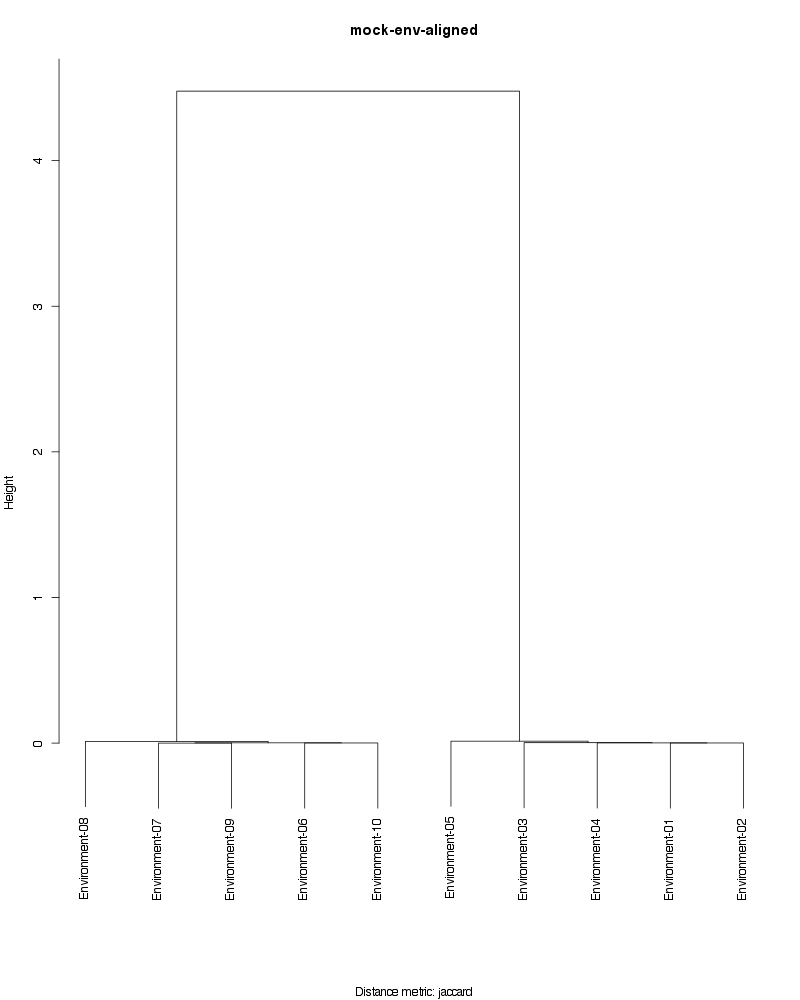
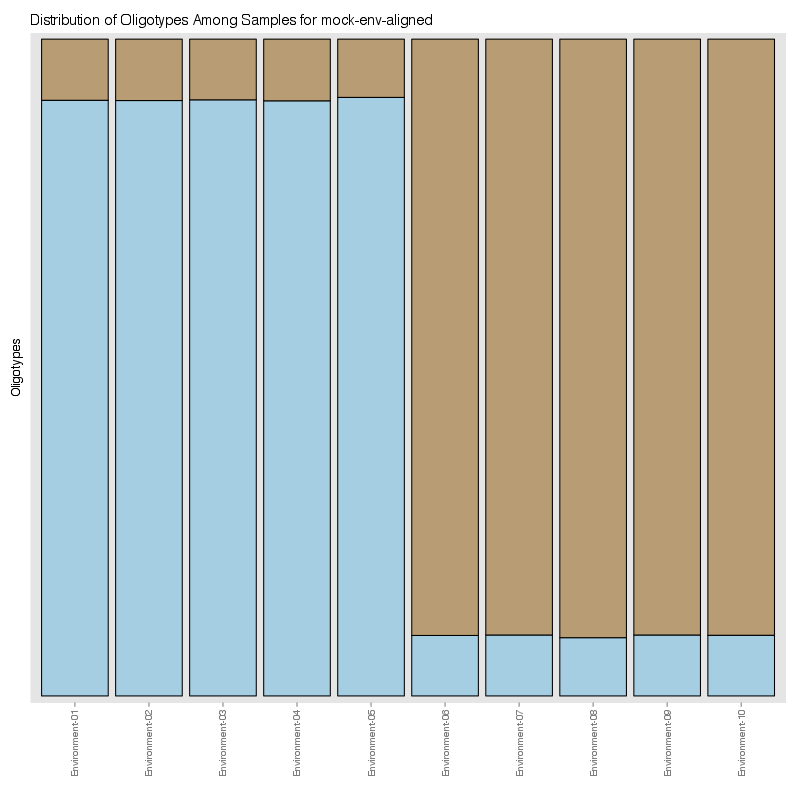
» Figure shows the oligotype distribution profiles among samples. TAB separated files matrix_percents.txt and matrix_counts.txt hold the information that were used to generate this figure.
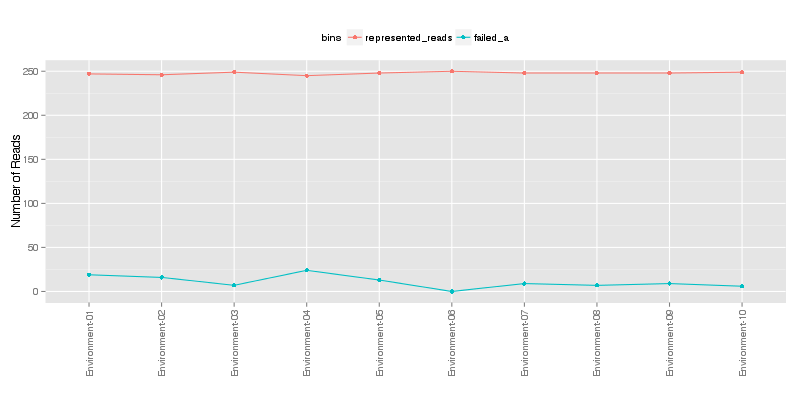
» Total number of reads for each sample that were analyzed.
» Abundant oligotypes along with their frequencies within the project are shown below. Every oligotype is followed by a representative sequence, which is the most frequent read in all reads that were collected by the given oligotype. Mouseover on an oligotype will popup a figure that shows the abundance distribution of unique sequences within the oligotype, along with the new entropy (in an ideal world there should be only one unique read with 0 entropy, but due to the random sequencing errors it is almost never the case). ✓ indicates that the most frequent sequence which was represented by the given oligotype hit something during the BLAST search with 100% identity in NCBI's nr database. In contrary, ✗ sign means that there was no perfect hit during the BLAST search.
GC b ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
✓ 1,243 GC b GT-TG-AAAAA-GTT-GGTG-GTG-AAAT-CCCAGA-GCTTAA-CT-CTGGAAC-TG-CCATC-AAAAC-TTTTCAG-CTA-GAGTAT-G-ATAGAGG-AAAGCAGAA-TTTCTAGT-G-TAGA-G-GTG-AAAT-TCG-T-AG-A-TATTA-G-AAA-GAATACCA-ATTG-CGA-AGGCAGC-TTTCT-GGATCATTACTGACA-C-TGAGGA-ACG-AAAGC-AT-GGGTAGCG-AAG-AGGA
✓ 1,235 AT b GT-TG-AAAAA-GTT-AGTG-GTG-AAAT-CCCAGA-GCTTAA-CT-CTGGAAC-TG-CCATT-AAAAC-TTTTCAG-CTA-GAGTAT-G-ATAGAGG-AAAGCAGAA-TTTCTAGT-G-TAGA-G-GTG-AAAT-TCG-T-AG-A-TATTA-G-AAA-GAATACCA-ATTG-CGA-AGGCAGC-TTTCT-GGATCATTACTGACA-C-TGAGGA-ACG-AAAGC-AT-GGGTAGCG-AAG-AGGA